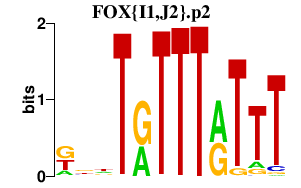
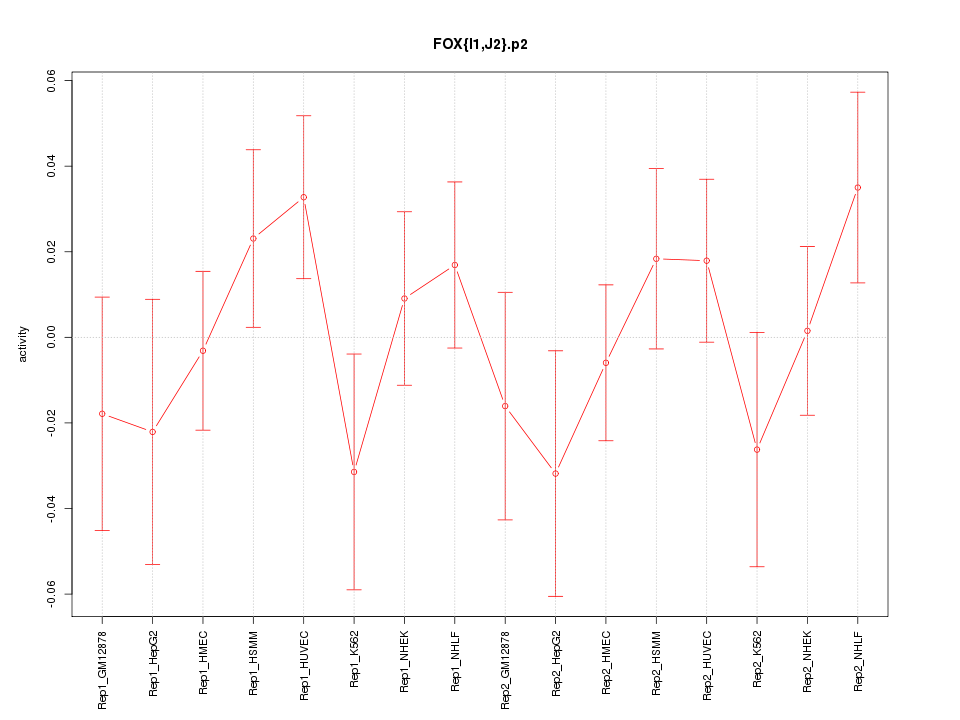
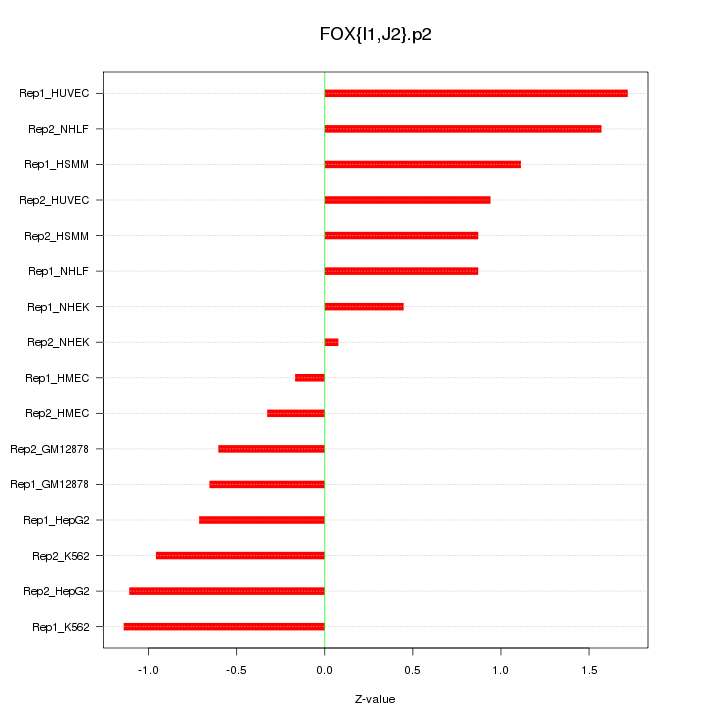
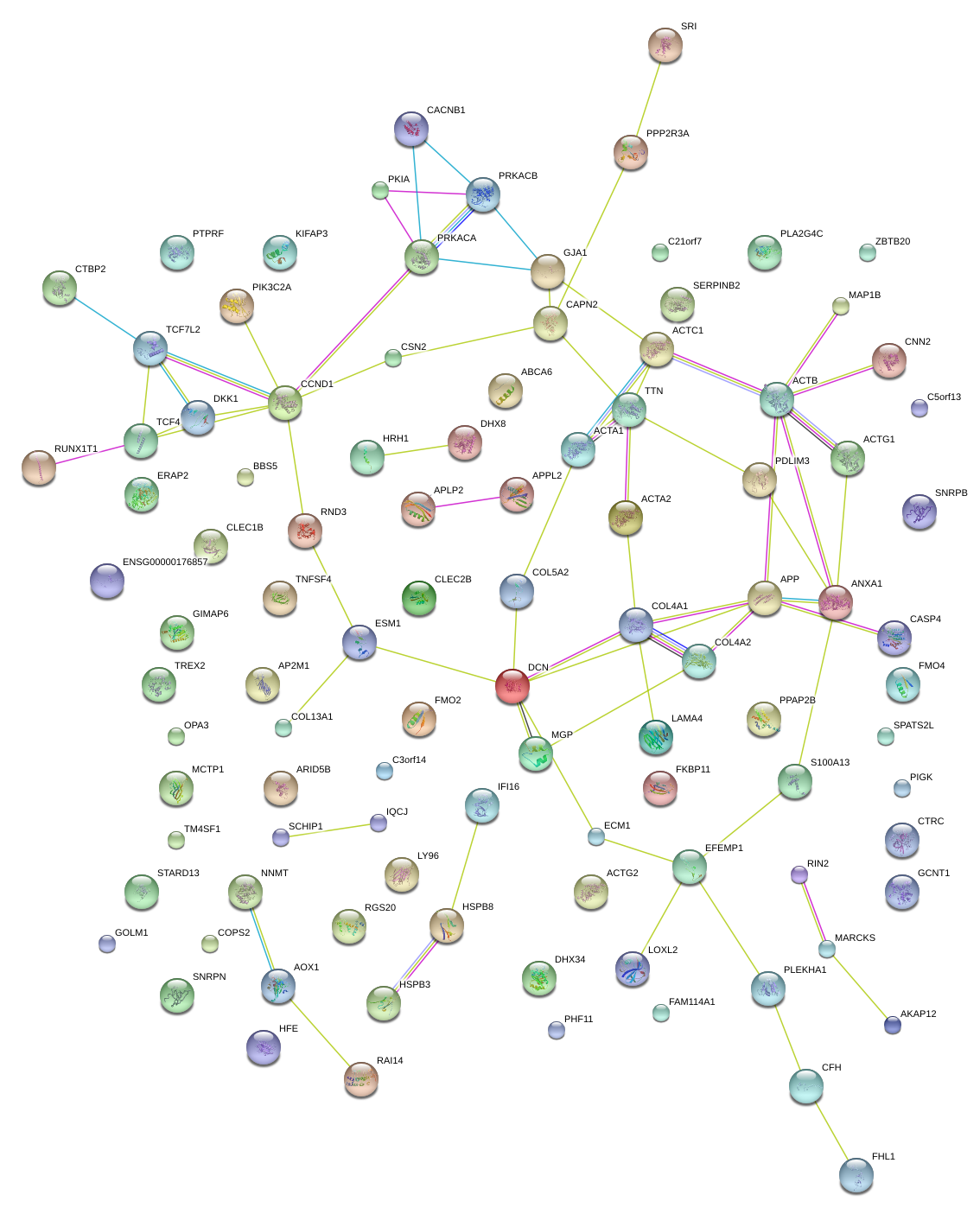
|
chr20_+_19870209
|
6.118
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_+_150480486
|
3.857
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr3_+_158787040
|
3.473
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr21_-_27512668
|
3.107
|
NM_001136016
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr20_+_19915716
|
2.946
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr9_+_75766777
|
2.434
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr13_+_50070096
|
2.319
|
|
PHF11
|
PHD finger protein 11
|
|
chr9_-_88715024
|
2.194
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr13_-_110802708
|
2.070
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr7_-_87849341
|
2.007
|
|
SRI
|
sorcin
|
|
chr2_-_190044330
|
1.919
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_-_149086884
|
1.829
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chrX_+_135278912
|
1.761
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_121756744
|
1.737
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr11_+_114167168
|
1.577
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr13_+_111134933
|
1.555
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr3_+_62305395
|
1.554
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr1_+_84609941
|
1.530
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr11_+_69467210
|
1.509
|
|
CCND1
|
cyclin D1
|
|
chr11_+_114167197
|
1.489
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr9_+_75766794
|
1.462
|
|
ANXA1
|
annexin A1
|
|
chr1_-_153599523
|
1.458
|
NM_001024213
|
S100A13
|
S100 calcium binding protein A13
|
|
chr1_+_158979681
|
1.408
|
NM_001206567
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr1_-_57045235
|
1.406
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_+_71561687
|
1.353
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr3_+_135741576
|
1.343
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr10_-_90712510
|
1.305
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr5_-_111312522
|
1.273
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr21_-_27253197
|
1.259
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr5_+_53751430
|
1.231
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr12_-_91576805
|
1.223
|
NM_001920
|
DCN
|
decorin
|
|
chr18_-_53303129
|
1.208
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr12_-_91576579
|
1.158
|
|
DCN
|
decorin
|
|
chr12_-_10021932
|
1.099
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr6_+_151646634
|
1.092
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_-_48614013
|
1.092
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr4_+_38869346
|
1.083
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr8_+_74903563
|
1.066
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr7_-_87849366
|
1.058
|
NM_003130
|
SRI
|
sorcin
|
|
chr5_-_54281345
|
1.048
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr6_-_112575766
|
1.038
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr6_-_112575880
|
1.036
|
|
LAMA4
|
laminin, alpha 4
|
|
chr13_-_33859835
|
1.023
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_+_96212167
|
1.015
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr11_-_104827421
|
1.000
|
NM_033306
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr1_+_171154387
|
0.999
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr5_-_94224601
|
0.975
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr2_-_56151273
|
0.970
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr1_+_84630575
|
0.964
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr21_+_30517897
|
0.964
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr2_+_201170603
|
0.961
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr12_-_15038724
|
0.961
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr10_+_71561643
|
0.960
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr18_-_53177999
|
0.959
|
NM_001243231
|
TCF4
|
transcription factor 4
|
|
chr15_+_25068793
|
0.950
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr10_-_126849066
|
0.945
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr10_+_124145584
|
0.941
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr2_+_170335948
|
0.934
|
NM_152384
|
BBS5
|
Bardet-Biedl syndrome 5
|
|
chr3_+_11267666
|
0.933
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr1_-_173174611
|
0.922
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr22_-_43036606
|
0.907
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr8_+_79428335
|
0.898
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr6_+_29855349
|
0.894
|
|
HLA-H
|
major histocompatibility complex, class I, H (pseudogene)
|
|
chr5_+_34687663
|
0.892
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr12_-_105627182
|
0.891
|
NM_001251905
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr1_+_15764934
|
0.889
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr6_-_112575708
|
0.886
|
|
LAMA4
|
laminin, alpha 4
|
|
chr1_-_57045214
|
0.884
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_-_114343052
|
0.858
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_71502700
|
0.851
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr7_-_150329285
|
0.849
|
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr10_-_126849010
|
0.846
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr1_+_44064509
|
0.845
|
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr10_+_54074037
|
0.845
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr1_-_170043608
|
0.844
|
NM_001204514
NM_001204516
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr4_-_186456580
|
0.831
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr12_+_119616594
|
0.828
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr1_-_77685076
|
0.823
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr11_-_17191287
|
0.819
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr8_-_23261675
|
0.819
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr2_+_201473942
|
0.813
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr16_+_8715473
|
0.794
|
NM_024109
|
METTL22
|
methyltransferase like 22
|
|
chr10_+_63808968
|
0.792
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr8_-_23261632
|
0.788
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr19_+_1026271
|
0.762
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr13_+_50069745
|
0.757
|
NM_001040443
|
PHF11
|
PHD finger protein 11
|
|
chr6_+_114178516
|
0.751
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr4_-_70826712
|
0.745
|
NM_001891
|
CSN2
|
casein beta
|
|
chr8_-_93111915
|
0.743
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_84630374
|
0.727
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_+_201170795
|
0.722
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr2_-_179672056
|
0.718
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr18_+_61554938
|
0.709
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr3_+_183892668
|
0.709
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr12_-_10022457
|
0.708
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr1_+_223900061
|
0.700
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr9_+_79115548
|
0.689
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr2_-_151344145
|
0.689
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr5_-_111091947
|
0.679
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_46688362
|
0.671
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr19_-_4400470
|
0.669
|
NM_001199943
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr1_-_163172863
|
0.662
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr4_+_146403955
|
0.658
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr3_+_183892662
|
0.656
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr19_+_50031232
|
0.655
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr17_-_48264039
|
0.647
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr21_-_27543445
|
0.641
|
NM_001136131
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr3_+_183892647
|
0.631
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr11_+_63304272
|
0.626
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr4_+_47487230
|
0.623
|
NM_020453
|
ATP10D
|
ATPase, class V, type 10D
|
|
chr4_+_89299874
|
0.620
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr9_+_124086842
|
0.616
|
|
GSN
|
gelsolin
|
|
chr9_-_14086901
|
0.610
|
|
NFIB
|
nuclear factor I/B
|
|
chr9_-_117880411
|
0.610
|
NM_002160
|
TNC
|
tenascin C
|
|
chr4_+_15704572
|
0.610
|
NM_004334
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr2_+_152214105
|
0.609
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr1_-_57045129
|
0.605
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_+_125695787
|
0.594
|
NM_001146319
|
GRAMD3
|
GRAM domain containing 3
|
|
chr7_+_76026819
|
0.593
|
NM_007155
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr14_+_62164339
|
0.591
|
NM_001243084
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr1_-_244006471
|
0.590
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr11_-_8739398
|
0.586
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr13_-_52980628
|
0.581
|
NM_018676
NM_199263
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr11_+_5710816
|
0.579
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr1_+_157963062
|
0.579
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr12_+_10365415
|
0.578
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr8_+_144640476
|
0.577
|
NM_024736
|
GSDMD
|
gasdermin D
|
|
chr6_+_153019029
|
0.577
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr20_+_9288446
|
0.576
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chrX_-_10851808
|
0.576
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr18_-_21852189
|
0.568
|
NM_018030
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr9_+_132962871
|
0.559
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr3_+_183892569
|
0.558
|
NM_001025205
NM_004068
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr15_+_25101697
|
0.557
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr4_-_16900028
|
0.557
|
|
LDB2
|
LIM domain binding 2
|
|
chr8_+_54764367
|
0.557
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr21_-_43346780
|
0.552
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr6_-_128841432
|
0.550
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr4_+_89299980
|
0.547
|
|
HERC6
|
hect domain and RLD 6
|
|
chr1_-_110933683
|
0.545
|
NM_001201546
NM_001201547
NM_001201548
NM_001201549
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr4_-_102268626
|
0.543
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr8_-_93107442
|
0.543
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_150122415
|
0.537
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr4_-_16900256
|
0.530
|
|
LDB2
|
LIM domain binding 2
|
|
chr3_+_45067750
|
0.524
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr2_+_102953716
|
0.522
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr1_+_171060017
|
0.518
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr15_+_64680002
|
0.515
|
NM_016213
|
TRIP4
|
thyroid hormone receptor interactor 4
|
|
chr18_+_72922725
|
0.515
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr4_+_77953942
|
0.513
|
|
SEPT11
|
septin 11
|
|
chr8_-_13134028
|
0.513
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_19969722
|
0.512
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr10_-_29754478
|
0.509
|
|
SVIL
|
supervillin
|
|
chr2_+_207406815
|
0.505
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr2_+_20646831
|
0.502
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr8_-_17658385
|
0.499
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chrX_+_114827818
|
0.495
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr5_+_140718353
|
0.489
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr19_-_52255099
|
0.489
|
NM_001193306
NM_002029
|
FPR1
|
formyl peptide receptor 1
|
|
chr4_-_102267952
|
0.489
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr4_-_16900348
|
0.486
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr22_-_36424397
|
0.478
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr12_-_102455763
|
0.476
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr7_-_93519990
|
0.476
|
NM_006528
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chr1_-_153599743
|
0.474
|
NM_001024212
|
S100A13
|
S100 calcium binding protein A13
|
|
chr2_-_188419185
|
0.471
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_-_227664330
|
0.471
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr8_-_93107881
|
0.462
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_110933635
|
0.462
|
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr13_-_33780142
|
0.461
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_128968470
|
0.459
|
|
COPG
|
coatomer protein complex, subunit gamma
|
|
chr12_+_56915587
|
0.457
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr7_+_89783688
|
0.456
|
NM_012449
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chr19_+_14491920
|
0.456
|
NM_001025160
NM_001784
NM_078481
|
CD97
|
CD97 molecule
|
|
chr3_-_15382874
|
0.455
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr9_-_16727887
|
0.453
|
|
BNC2
|
basonuclin 2
|
|
chr3_-_114102488
|
0.450
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr18_+_22006611
|
0.449
|
|
IMPACT
|
Impact homolog (mouse)
|
|
chr12_+_78225068
|
0.449
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr6_-_134496833
|
0.443
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_140734767
|
0.441
|
NM_018917
NM_032053
|
PCDHGA4
|
protocadherin gamma subfamily A, 4
|
|
chr4_+_90816002
|
0.439
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr8_-_12973752
|
0.439
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr20_-_44538654
|
0.438
|
|
PLTP
|
phospholipid transfer protein
|
|
chr5_-_68616299
|
0.437
|
NM_176816
|
CCDC125
|
coiled-coil domain containing 125
|
|
chr1_+_84630014
|
0.437
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr19_-_4400357
|
0.436
|
NM_001199944
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr1_+_82266081
|
0.435
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr7_+_89783789
|
0.434
|
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chrX_+_80457302
|
0.434
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chrX_-_21776158
|
0.434
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr2_-_188312872
|
0.431
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr1_+_101185195
|
0.428
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr2_-_133429069
|
0.428
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr3_+_35721115
|
0.426
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_-_95064213
|
0.426
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr8_-_108510094
|
0.421
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr20_+_9198036
|
0.421
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chrX_-_6145887
|
0.421
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr12_-_102455793
|
0.420
|
NM_016053
|
CCDC53
|
coiled-coil domain containing 53
|